Hierarchical Clustering for Categorical and Mixed Data Types in Python
Hierarchical clustering is one of the most popular clustering algorithms after partitioning clustering algorithms like k-means clustering. In this article, we will discuss hierarchical clustering for categorical and mixed data types in python. For this, we will implement agglomerative clustering for datasets having categorical data and mixed data types.
How to Perform Hierarchical Clustering for Categorical and Mixed Data Types?
Most of the clustering algorithms are primarily defined for numeric data types. However, we can perform clustering on categorical data or a dataset having mixed data types if we define the function to calculate the distance between the data points in such a dataset.
In hierarchical clustering, we need to create a linkage matrix from the distance matrix of the dataset. So, if we define a function to calculate the distance between categorical or mixed data types, we can implement agglomerative clustering on the dataset.
To implement hierarchical clustering on categorical or mixed data types, you can use the functions defined in this article on silhouette coefficient for k-modes and k-prototypes clustering.
Hierarchical Clustering for Categorical Data in Python
To implement agglomerative hierarchical clustering on categorical data, we will use the create_dm() function defined in the above-mentioned article to calculate the distance matrix for the given dataset.
- We will determine the distance matrix by using the dissimilarity score between two pieces of data. In a previous article on k-modes clustering with numerical examples, I explained how to calculate dissimilarity scores for categorical data and provided a numerical example.
- To implement the calculation of the dissimilarity score in python, we will utilize the
kprototypes.matching_dissim() function. The operation of this function is covered in an article on clustering mixed data types in Python. - Using the
matching_dissim()function, we implemented thecreate_dm()function which is discussed in the article on silhouette coefficient. - Using the
create_dm()function, we will first calculate the distance matrix for a given dataset having categorical data. - Next, we will pass the distance matrix to the
linkage()function defined in thescipy.cluster.hierarchymodule. Thelinkage()function will return a linkage matrix after execution.
Once we get the linkage matrix, we can plot the dendrogram for the given categorical data. I have already discussed how to plot a dendrogram in python. You can read this article to understand how to plot a dendrogram.
import pandas as pd
from scipy.spatial import distance_matrix
from scipy.cluster.hierarchy import dendrogram, linkage
import matplotlib.pyplot as plt
import numpy as np
def create_dm(dataset):
#if the input dataset is a dataframe, we take out the values as a numpy.
#If the input dataset is a numpy array, we use it as is.
if type(dataset).__name__=='DataFrame':
dataset=dataset.values
lenDataset=len(dataset)
distance_matrix=np.zeros(lenDataset*lenDataset).reshape(lenDataset,lenDataset)
for i in range(lenDataset):
for j in range(lenDataset):
x1= dataset[i].reshape(1,-1)
x2= dataset[j].reshape(1,-1)
distance=kprototypes.matching_dissim(x1, x2)
distance_matrix[i][j]=distance
distance_matrix[j][i]=distance
return distance_matrix
data=pd.read_csv("KModes-dataset.csv", index_col=["Student"])
distance_matrix=create_dm(data)
linkage_matrix = linkage(distance_matrix, "ward")
dendrogram(linkage_matrix, labels=data.index)
plt.title("Dendrogram Using ward Linkage")
plt.xticks(rotation='vertical')
plt.show()Dataset:
Output:
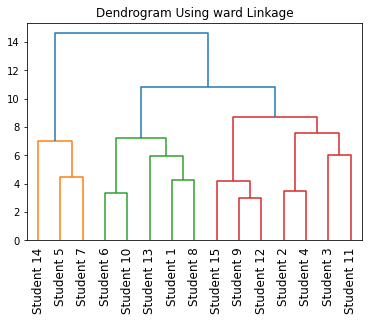
In the above code, we have plotted a dendrogram for categorical data using the scipy module.
Instead of plotting the dendrogram, you can also find cluster labels of different clusters by performing hierarchical agglomerative clustering on categorical data as shown below.
import pandas as pd
from scipy.spatial import distance_matrix
from scipy.cluster.hierarchy import dendrogram, linkage, fcluster
import matplotlib.pyplot as plt
import numpy as np
def create_dm(dataset):
#if the input dataset is a dataframe, we take out the values as a numpy.
#If the input dataset is a numpy array, we use it as is.
if type(dataset).__name__=='DataFrame':
dataset=dataset.values
lenDataset=len(dataset)
distance_matrix=np.zeros(lenDataset*lenDataset).reshape(lenDataset,lenDataset)
for i in range(lenDataset):
for j in range(lenDataset):
x1= dataset[i].reshape(1,-1)
x2= dataset[j].reshape(1,-1)
distance=kprototypes.matching_dissim(x1, x2)
distance_matrix[i][j]=distance
distance_matrix[j][i]=distance
return distance_matrix
data=pd.read_csv("KModes-dataset.csv", index_col=["Student"])
distance_matrix=create_dm(data)
linkage_matrix = linkage(distance_matrix, "ward")
cluster_labels = fcluster(linkage_matrix,3,criterion='maxclust')
data["Cluster"]=cluster_labels
print("The clustered data is:")
print(data)Output:
The clustered data is:
Subject 1 Subject 2 Subject 3 Subject 4 Subject 5 Cluster
Student
Student 1 A B A B A 2
Student 2 A A B B A 3
Student 3 C C B A C 3
Student 4 C A B B A 3
Student 5 B A A B C 1
Student 6 A B B A C 2
Student 7 B A C C C 1
Student 8 A A A A A 2
Student 9 A C B B B 3
Student 10 A B B A A 2
Student 11 C C D B A 3
Student 12 A C B B C 3
Student 13 A B A C B 2
Student 14 B C C D B 1
Student 15 A B B B B 3Instead of using the scipy module and calculating the linkage matrix, you can directly implement hierarchical clustering on categorical data using the sklearn module in python as shown below.
from sklearn.cluster import AgglomerativeClustering
import pandas as pd
import numpy as np
def create_dm(dataset):
#if the input dataset is a dataframe, we take out the values as a numpy.
#If the input dataset is a numpy array, we use it as is.
if type(dataset).__name__=='DataFrame':
dataset=dataset.values
lenDataset=len(dataset)
distance_matrix=np.zeros(lenDataset*lenDataset).reshape(lenDataset,lenDataset)
for i in range(lenDataset):
for j in range(lenDataset):
x1= dataset[i].reshape(1,-1)
x2= dataset[j].reshape(1,-1)
distance=kprototypes.matching_dissim(x1, x2)
distance_matrix[i][j]=distance
distance_matrix[j][i]=distance
return distance_matrix
data=pd.read_csv("KModes-dataset.csv", index_col=["Student"])
distance_matrix=create_dm(data)
model = AgglomerativeClustering(n_clusters=3, affinity="precomputed", linkage='complete')
model.fit(distance_matrix)
labels = model.labels_
data["Cluster"]=labels
print("The clustered data is:")
print(data)Output:
The clustered data is:
Subject 1 Subject 2 Subject 3 Subject 4 Subject 5 Cluster
Student
Student 1 A B A B A 0
Student 2 A A B B A 0
Student 3 C C B A C 0
Student 4 C A B B A 0
Student 5 B A A B C 2
Student 6 A B B A C 1
Student 7 B A C C C 2
Student 8 A A A A A 0
Student 9 A C B B B 0
Student 10 A B B A A 1
Student 11 C C D B A 0
Student 12 A C B B C 0
Student 13 A B A C B 1
Student 14 B C C D B 2
Student 15 A B B B B 1In the above example, we have used the sklearn module for hierarchical clustering for categorical data. Here, we didn’t need to calculate the linkage matrix. The AgglomerativeClustering() function directly takes the distance matrix as its input argument. Also, you should set the affinity parameter to "precomputed" if you are using sklearn version 1.4 or earlier. For versions above this, you should set the metric parameter to precomputed.
Hierarchical Clustering for Mixed Data Types in Python
By calculating the distance matrix, you can also implement agglomerative hierarchical clustering for mixed data types in python. For this, we will use the following steps.
- First, we will define a function to calculate the distance between two data points having mixed attributes. For this, we will calculate the dissimilarity between categorical values and the distance between the numeric attributes separately. I have already implemented this in the
mixed_distance()function in this article. - After defining the function to calculate the distance between the data points, you can define a function to calculate the distance matrix for the given dataset with attributes having mixed data types. I have implemented the
dm_prototypes()function to calculate the same in the same article on silhouette coefficient. - After calculating the distance matrix for the dataset having mixed data types, we will pass the distance matrix to the
linkage()function defined in thescipy.cluster.hierarchymodule to create a linkage matrix. Thelinkage()function will return a linkage matrix after execution.
Once we get the linkage matrix, we can plot the dendrogram for the given dataset having mixed data types using the dendrogram() function as shown below.
import pandas as pd
from scipy.spatial import distance_matrix
from scipy.cluster.hierarchy import dendrogram, linkage, fcluster
import matplotlib.pyplot as plt
import numpy as np
def mixed_distance(a,b,categorical=None, alpha=0.01):
if categorical is None:
num_score=kprototypes.euclidean_dissim(a,b)
return num_score
else:
cat_index=categorical
a_cat=[]
b_cat=[]
for index in cat_index:
a_cat.append(a[index])
b_cat.append(b[index])
a_num=[]
b_num=[]
l=len(a)
for index in range(l):
if index not in cat_index:
a_num.append(a[index])
b_num.append(b[index])
a_cat=np.array(a_cat).reshape(1,-1)
a_num=np.array(a_num).reshape(1,-1)
b_cat=np.array(b_cat).reshape(1,-1)
b_num=np.array(b_num).reshape(1,-1)
cat_score=kprototypes.matching_dissim(a_cat,b_cat)
num_score=kprototypes.euclidean_dissim(a_num,b_num)
return cat_score+num_score*alpha
def dm_prototypes(dataset,categorical=None,alpha=0.1):
#if the input dataset is a dataframe, we take out the values as a numpy.
#If the input dataset is a numpy array, we use it as is.
if type(dataset).__name__=='DataFrame':
dataset=dataset.values
lenDataset=len(dataset)
distance_matrix=np.zeros(lenDataset*lenDataset).reshape(lenDataset,lenDataset)
for i in range(lenDataset):
for j in range(lenDataset):
x1= dataset[i]
x2= dataset[j]
distance=mixed_distance(x1, x2,categorical=categorical,alpha=alpha)
distance_matrix[i][j]=distance
distance_matrix[j][i]=distance
return distance_matrix
import pandas as pd
import numpy as np
df=pd.read_csv("Kprototypes_dataset.csv", index_col=["Student"])
#Normalize dataset
df["Height(in cms)"]=(df["Height(in cms)"]/df["Height(in cms)"].abs().max())*5
df["Weight(in Kgs)"]=(df["Weight(in Kgs)"]/df["Weight(in Kgs)"].abs().max())*5
#obtain array of values
data_array=df.values
#specify data types
data_array[:, 0:3] = data_array[:, 0:3].astype(str)
data_array[:, 3:] = data_array[:, 3::].astype(float)
distance_matrix=dm_prototypes(data_array,categorical=[0, 1, 2],alpha=0.1)
linkage_matrix = linkage(distance_matrix, "ward")
dendrogram(linkage_matrix, labels=data.index)
plt.title("Dendrogram Using ward Linkage")
plt.xticks(rotation='vertical')
plt.show()Dataset:
Output:
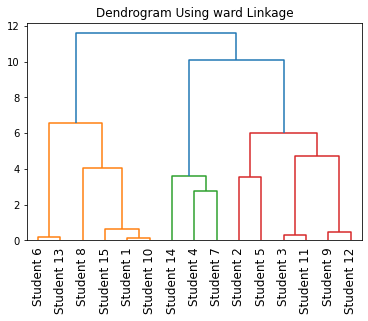
Instead of plotting the dendrogram, you can also find cluster labels of different clusters by performing hierarchical agglomerative clustering on mixed data types as shown below.
import pandas as pd
from scipy.spatial import distance_matrix
from scipy.cluster.hierarchy import dendrogram, linkage, fcluster
import matplotlib.pyplot as plt
import numpy as np
def mixed_distance(a,b,categorical=None, alpha=0.01):
if categorical is None:
num_score=kprototypes.euclidean_dissim(a,b)
return num_score
else:
cat_index=categorical
a_cat=[]
b_cat=[]
for index in cat_index:
a_cat.append(a[index])
b_cat.append(b[index])
a_num=[]
b_num=[]
l=len(a)
for index in range(l):
if index not in cat_index:
a_num.append(a[index])
b_num.append(b[index])
a_cat=np.array(a_cat).reshape(1,-1)
a_num=np.array(a_num).reshape(1,-1)
b_cat=np.array(b_cat).reshape(1,-1)
b_num=np.array(b_num).reshape(1,-1)
cat_score=kprototypes.matching_dissim(a_cat,b_cat)
num_score=kprototypes.euclidean_dissim(a_num,b_num)
return cat_score+num_score*alpha
def dm_prototypes(dataset,categorical=None,alpha=0.1):
#if the input dataset is a dataframe, we take out the values as a numpy.
#If the input dataset is a numpy array, we use it as is.
if type(dataset).__name__=='DataFrame':
dataset=dataset.values
lenDataset=len(dataset)
distance_matrix=np.zeros(lenDataset*lenDataset).reshape(lenDataset,lenDataset)
for i in range(lenDataset):
for j in range(lenDataset):
x1= dataset[i]
x2= dataset[j]
distance=mixed_distance(x1, x2,categorical=categorical,alpha=alpha)
distance_matrix[i][j]=distance
distance_matrix[j][i]=distance
return distance_matrix
import pandas as pd
import numpy as np
df=pd.read_csv("Kprototypes_dataset.csv", index_col=["Student"])
#Normalize dataset
df["Height(in cms)"]=(df["Height(in cms)"]/df["Height(in cms)"].abs().max())*5
df["Weight(in Kgs)"]=(df["Weight(in Kgs)"]/df["Weight(in Kgs)"].abs().max())*5
#obtain array of values
data_array=df.values
#specify data types
data_array[:, 0:3] = data_array[:, 0:3].astype(str)
data_array[:, 3:] = data_array[:, 3::].astype(float)
distance_matrix=dm_prototypes(data_array,categorical=[0, 1, 2],alpha=0.1)
linkage_matrix = linkage(distance_matrix, "ward")
cluster_labels = fcluster(linkage_matrix,3,criterion='maxclust')
#reading dataframe again as featurs were normalized earlier
df=pd.read_csv("Kprototypes_dataset.csv", index_col=["Student"])
df.columns=["IQ", "EQ", "Gender", "Height", "Weight"]
df["Cluster"]=cluster_labels
print("The clustered data is:")
print(df)Output:
The clustered data is:
IQ EQ Gender Height Weight Cluster
Student
Student 1 A B F 155 53 1
Student 2 A A M 174 70 3
Student 3 C C M 177 75 3
Student 4 C A F 182 80 2
Student 5 B A M 152 76 3
Student 6 A B M 160 69 1
Student 7 B A F 175 55 2
Student 8 A A F 181 70 1
Student 9 A C M 180 85 3
Student 10 A B F 166 54 1
Student 11 C C M 162 66 3
Student 12 A C M 153 74 3
Student 13 A B M 160 62 1
Student 14 B C F 169 59 2
Student 15 A B F 171 71 1Instead of using the scipy module and calculating the linkage matrix, you can directly implement hierarchical clustering using the sklearn module on mixed data types in python as shown below.
from sklearn.cluster import AgglomerativeClustering
import pandas as pd
import numpy as np
def mixed_distance(a,b,categorical=None, alpha=0.01):
if categorical is None:
num_score=kprototypes.euclidean_dissim(a,b)
return num_score
else:
cat_index=categorical
a_cat=[]
b_cat=[]
for index in cat_index:
a_cat.append(a[index])
b_cat.append(b[index])
a_num=[]
b_num=[]
l=len(a)
for index in range(l):
if index not in cat_index:
a_num.append(a[index])
b_num.append(b[index])
a_cat=np.array(a_cat).reshape(1,-1)
a_num=np.array(a_num).reshape(1,-1)
b_cat=np.array(b_cat).reshape(1,-1)
b_num=np.array(b_num).reshape(1,-1)
cat_score=kprototypes.matching_dissim(a_cat,b_cat)
num_score=kprototypes.euclidean_dissim(a_num,b_num)
return cat_score+num_score*alpha
def dm_prototypes(dataset,categorical=None,alpha=0.1):
#if the input dataset is a dataframe, we take out the values as a numpy.
#If the input dataset is a numpy array, we use it as is.
if type(dataset).__name__=='DataFrame':
dataset=dataset.values
lenDataset=len(dataset)
distance_matrix=np.zeros(lenDataset*lenDataset).reshape(lenDataset,lenDataset)
for i in range(lenDataset):
for j in range(lenDataset):
x1= dataset[i]
x2= dataset[j]
distance=mixed_distance(x1, x2,categorical=categorical,alpha=alpha)
distance_matrix[i][j]=distance
distance_matrix[j][i]=distance
return distance_matrix
import pandas as pd
import numpy as np
df=pd.read_csv("Kprototypes_dataset.csv", index_col=["Student"])
#Normalize dataset
df["Height(in cms)"]=(df["Height(in cms)"]/df["Height(in cms)"].abs().max())*5
df["Weight(in Kgs)"]=(df["Weight(in Kgs)"]/df["Weight(in Kgs)"].abs().max())*5
#obtain array of values
data_array=df.values
#specify data types
data_array[:, 0:3] = data_array[:, 0:3].astype(str)
data_array[:, 3:] = data_array[:, 3::].astype(float)
distance_matrix=dm_prototypes(data_array,categorical=[0, 1, 2],alpha=0.1)
distance_matrix=create_dm(data)
model = AgglomerativeClustering(n_clusters=3, affinity="precomputed", linkage='complete')
model.fit(distance_matrix)
labels = model.labels_
df=pd.read_csv("Kprototypes_dataset.csv", index_col=["Student"])
df.columns=["IQ", "EQ", "Gender", "Height", "Weight"]
df["Cluster"]=labels
print("The clustered data is:")
print(df)Output:
The clustered data is:
IQ EQ Gender Height Weight Cluster
Student
Student 1 A B F 155 53 0
Student 2 A A M 174 70 0
Student 3 C C M 177 75 0
Student 4 C A F 182 80 0
Student 5 B A M 152 76 2
Student 6 A B M 160 69 1
Student 7 B A F 175 55 2
Student 8 A A F 181 70 0
Student 9 A C M 180 85 0
Student 10 A B F 166 54 1
Student 11 C C M 162 66 0
Student 12 A C M 153 74 0
Student 13 A B M 160 62 1
Student 14 B C F 169 59 2
Student 15 A B F 171 71 1Conclusion
In this article, we discussed how to perform hierarchical agglomerative clustering on categorical and mixed data types. We also discussed how to plot dendrograms for categorical and mixed data types. For this, we used the scipy and sklearn modules in python.
To learn more about machine learning, you can read this article on regression in machine learning. You might also like this article on polynomial regression using sklearn in python.
I hope you enjoyed reading this article. Stay tuned for more informative articles.
Happy Learning!
